Selected thesis
Find here some selected completed thesis and reserach projects in the following fields:
- Machine learning and deep neural networks
- Acausal modelling of biological systems
- Drug design and molecular modelling
- Development informatics for clinical studies
 | Julia package: NNHelferlein.jl |
2022, Project
Collection of little helpers to simplify various Machine Learning tasks such as designing neural networks in Julia. The high-level API makes its possible to define and train neural networks in less lines of code and even more intuitively as with high-level frameworks while keeping the full control over all implementation details.
NNHelferlein at GitHub |
 | U-Net based Gland and Nuclei Segmentation for the Classification of Gleason Patterns in Biopsy whole slide image (WSI) patches |
2021, Master's Project
In gleason grading, tumor patterns are differentiated based on the morphology of prostate glands and the distribution of nuclei. This study provides an image segmentation approach usable for ML based gleason grading that fully relies on gland morphology and nuclei distribution.
|
 | R-package: RHRT |
2021, Project
Methods to scan RR interval data for Premature Ventricular Complexes (PVCs) and parameterise and plot the resulting Heart Rate Turbulence (HRT).
RHRT at CRAN |
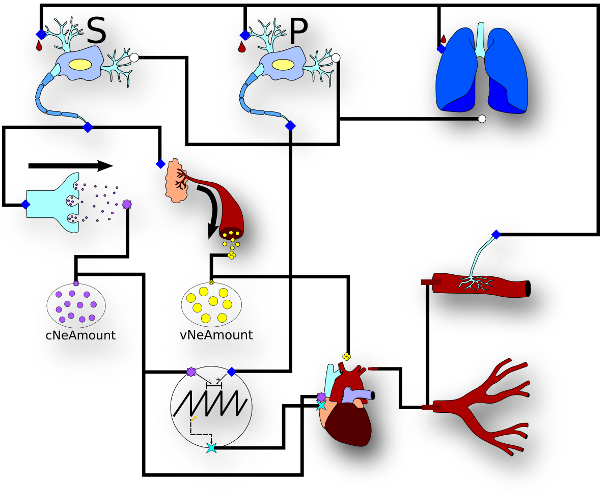 | Modelica model of the human baroreflex. |
2021, Project
Implementation of a heretogeneous multi-scale model of the human baroreflex in Modelica following the MoDROGH criteria: Modular, Descriptive, human-Readable, Open, Graphical and Hybrid.
J. Sys. Biol. paper |
 | Julia-package for analysis of heart-rate variability and classification of ECGs by combining recurrence plots and deep neural networks. |
2020, Bachelor's Project
This package implements the most common methods for heart rate variability analysis.
HeartRateVariability.jl at GitHub |
 | Simulation of the blood coagulation cascade and its dependency on vitamin K synthesis and warfarin. |
2020, Bachelor's Project
Modelica-based simulation of warfarin-dependent blood coagulation including pharmacokinetics of oral, IM- or IV-application of warfarin.
|
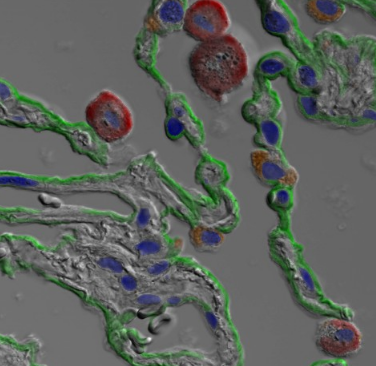 | A deep learning approach towards automated cell type labeling in human lung tissue. |
2020, Master's Project
A deep neural network is developed to simulate and predict fluorescent stainings from unstained microscopic images. Additionally, visualization of the predictions are generated to enable the application in the biomedical field.
|
 | Deep neural architecture for ECG anomaly detection |
2019, Bachelor's Project
An unsupervised DNN is developed to adapt to patient-individual ECG characteristics and recognise critical conditions.
|
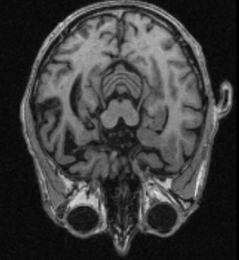 | Diagnosis of Alzheimer′s disease with help of deep learning |
2018, Bachelor′s thesis
Novel deep learning architectures are developed to handle 3D MRT data and to recognise differences between deseased and a healthy control group.
|
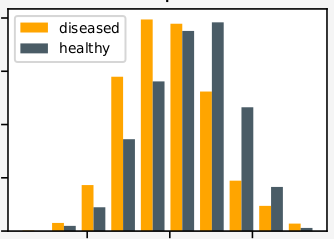 | Interpretation of HRV time-series by mulit-fractal analysis and deep learning. |
2018, Bachelor′s Project
A transfer learning approach is applied for training a deep neural architecture in order to overcome the limitation of inconsitent clinical data in the field of ECG analysis.
|
 | A Julia package for training and visualisation of Kohonen′s self-organising maps. |
2018, Project
The package provides training and visualisation functions for rectangular, hexagonal and spherical SOMs. Training functions are implemented in pure Julia. Project on GitHub:
LiScI-Lab/SOM.jl |
 | Proposal and implementaion of a lightweight development environment for the acausal, object-oriented modelling language Modelica. |
2017, several projects and thesis
A bunch of lightweight tools are developed to facilitate model-development with OpenModelica. Details can be found in the corresponding GitHub project:
GitHub project of MoTE |
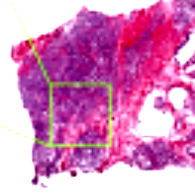 | Segmentation and classification of histopathological slides |
2017, Bachelor's Thesis
Patterns of cell nuclei in H&E-stained histopathological slides are identified and matched to according Gleason score by mapping to self organising maps.
|
 | High-throughput Software Pipelines for Genotypic Antimicrobial Resistance Testing |
2016, Thesis
A pipeline for sequence-based resistance testing is developed and applied to multi-resistant hospital pathogen S. Aureus. This technique may bring benefits in terms of cost- and time-effectiveness compared to previously established methods.
|
 | Acausal modelling of the cell cycle |
2016, Master's Project
Regulatory pathways affecting the cell cycle have been implemented in the object-oriented modelling language Modelica. The simulations are integrated with a simulation of tumour growth.
|
 | Using Deep Gated RNN with a Convolutional Front End for End-to-End Classification of Heart Sound |
2016, Master's Project
Classification of heart sounds (phonocardiograms, PCGs) is the challenging objective of the 2016 PhysioNet Challenge. Our architecture combines convolutional and recurrent layers, followed by an attention mechanism, which weights time steps by importance and a dense multilayer perceptron as classifier.
|
 | Clinical *omics data handling in oncology |
2015, Thesis
An integrated system of R S4-classes is implemented for management, processing and visualisation of patient centred high-throughput sequencing data. Results are applied to clinical development projetcs in oncology.
|
 | Off-target-activity prediction for drug molecules |
2015, Thesis
An novel algorithm is developed to identify off-target activities of small molecules drugs.
|
 | Silicon Heart - An easy to use interactive real-time baroreflex simulator |
2015, Project
A simulator of the baroreflex loop is implemented as a distributed system, including independent functional units, with each of them running without synchronisation and in real-time.
|
 | 3-dimensional reconstruction of tumours from MRI volume data |
2015, Thesis
An algorithm is developed and applied to 3-dimensional reconstruction of tumours from MRI data. In addition, tissue segmentation and recognition of tumour boundaries are implemented.
|
 | In-silico assessment of molecular antibody properties |
2014, Thesis
The developed method allows the sequence-based prediction of mAB properties and supports candidate selection during the development of therapeutic antibodies.
|
 | Enhancements of a model for heart-rate variability |
2014, Thesis
A well established model for heart-rate variability has been enhanced by adding additional feedback loops. Analysis of the resulting HRV simulations demonstrate the realistic behaviour of the model.
|
 | Analysis of heart-rate variability |
2014, Thesis
HRV of several clinical datasets are analysed by means of their fractal properties. An easy-to-use web-based analysis pipeline is implementend.
|
 | Classification of biosignals with Learning Vecor Quantisation |
2014, Thesis
Kohonen's LVQ algorithms were implemented as VIs in the LabVIEW programming language G and applied to identify biosignals. An example application have been trained to recognise QRS complexes in heavily interfered ECGs.
|
 | Development of a novel algorithm for segmentation of histopathological slides |
2014, Thesis
An optimised algorithm, based on density-based clustering, is developped and applied to identification of nuclei in H&E-stained histopathological slides.
Project page will be available soon! |
 | Optimisation of a de-novo assembly pipeline |
2013, Thesis
A novel pipeline was developped to optimise the de-novo assembly of short fragments from next-generation sequencing data.
|
 | Development of a novel algorithm for detection and correction of frameshifts |
2013, Thesis
The multi-step algorithms detects frameshifts in de-novo transcriptom sequencing data. Mass spectroscopy data and blastx alignments are used to to create a high quality data set for training of a Hidden-Markov Model.
|
 | Development of KNIME nodes for accessing structured information from public and proprietary bioinformatics databases. |
2013, Project
Several KNIME nodes which allow querying heterogeneous bioinformatics information sources are developed in collaboration with an industry partner.
Project page will be available soon! |
 | Development of a portal for cardiovascular system simulation. |
2012, Thesis
A web portal for cardiovascular system simulation and a first simulation model have been implemented and applied for disease simulation.
|
 | Application of artificial neural networks to analysis and classification of medical data. |
2012, Thesis
Novel artificial neural network topologies are developed and applied to classification of gene expression data from lung cancer samples. The neural network predicts patient prognosis with higher accuracy compared to statistical methods or datamining algorithms.
|
 | Gene-expression analysis of a mouse MCAO Experiment |
2012, Thesis
Raw affymetrix gene-expression data of a MCAO experiment (middle cerebral arteria occlusion) without replicates are analysed by means of a hierarchical Bayes model. The method proved its ability to identify statistically significant differential gene expression from single experiment data.
|
 | Analysis of the influence of nucleotide variations on microarrays |
2012, Thesis
RNA-SEQ and microarray data are combined to study the influence of nucleotide variations on microarray experiments.
|
 | Target enrichment by array hybridisation |
2012, Thesis
The goal of this project is to develop a target enrichment method for the mus musculus whole genome based on DNA microarray capture and T7 linear amplification of cancer-related genes.
|
 | Development of a web-based knowledge-management portal |
2012, Thesis
Requirement specification, project management, design and implementation of a web-based knowledge-management portal, by means of model-driven software development.
|
 | In-silico prediction of liquid crystal properties |
2012, Project
Develpoment of methods and an intranet-based service for in-silico preciction of properties of materials for liquid cristals and OLEDs.
|
 | Bioinformatics KNIME nodes |
2011, Project
Several KNIME nodes for accessing bioinformatics data sources as well as for reading and writing of BiFx fileformats have been developed.
|
 | Representation of molecular structures as non-cyclic graphs |
2011, Thesis
Develpoment of a novel algorithm for describing chemical molecules as non-cyclic graphs. Several algorithms are compared and finally implemented as KNIME nodes.
|
 | Galaxy bioinformatics workflow management |
2011, P. Deuster
Galaxy is a web application, developed at Pennsylvania State University designed for analysis of genomics data. It provides a web-based user interface to bioinformatics tools, scripts and data analysis pipelines. Our Galaxy installation is linked to an Oracle Grid Engine (former SGE) to run bioinformatics tools parallelised on our HPC cluster. This project is a collaboration with the MPI Bad Nauheim. For the moment, the service is available only for internal use and for collaboration partners.
Visit the Galaxy home. |
 | Prediction of protonation in ligand-protein-complexes |
2011, B. Schulz
An efficient and reliable method for prediction of protonation in ligand-protein-complexes is developed. Tautomers, intra- and intermolecular hydrogen bonds are predicted and considered. An empirical scoring function is used for optimisation. The project was performed at the University of Hamburg, Center for Bioinformatics, with the group of Matthias Rarey.
|
 | Development and evaluation of a data analysis pipeline for MS-based quantitative phosphoproteomics |
2011, N. Schnell
A data analysis pipeline is developed to identify known and unknown phosphorylation sites. The pipeline is applied to compare example data from C2C12-cell culture and samples of Mus Muscula skeletal muscle. Technologies comprise SILAC, mass spectroscopy, Galaxy and scripting in R and Perl.
|
 | Fast generation of unambiguous molecular formula |
2011, B. Schulz
Implementation of a gold standard algorithm for generation of molecular 2-D-formula. The algorithm includes ring-peeling, a database of templates and a 2-D-forcefield. The project was performed at the University of Hamburg, Center for Bioinformatics, with the group of Matthias Rarey.
|
 | Development of a T7 based amplification for comparative genomic hybridization |
2011, J. Krauskopf
Development and evaluation of a whole genome amplification based on the T7 RNA polymerase which is suitable for array comparative genomic hybridization analysis and resequencing.
|
 | Oracle Grid Engine (former Sun Grid Engine) |
2011, M. Haub
Oracle Grid Engine software is a distributed resource management system that manages the distribution of users' workloads to available compute resources. In scope of the project, OGE (SGE) was set up as a front-end for our HPC cluster and parallelised bioinformatics tools, such as blast, have been implemented. The tools are integrated with a Galaxy workflow management system. For the moment, the grid is available only for internal use and for collaboration partners.
|
 | Development of a clinical gene expression database |
2011, Thesis
The database manages huge amounts of data from clinical gene expression experiments and integrates with other clinical parameters and biomarkers. It includes a front-end for searching and mining the data. This project is a collaboration with Michael Maier, JLU Giessen.
|
 | Development of an sncRNA Database |
2010, J. Pischimarov
The sRNAdb workbench aims to collect all published and predicted ncRNAs of Gram-positive bacteria and to make them available to the public for comparative analysis. Currently included are all available data of transcriptomic publications on this topic as well as SIPHT in-silico predictions.
Detailed information and web frontend can be found on the project page. |
 | New web-frontend for pdb-care |
2010, D. Mokros
The software detects and visualises discrepancies in connectivities and nomenclature of PDB entries containing carbohydrates. This project is a collaboration with Prof. Dr. Thomas Lütteke, JLU Giessen.
Visit the new PDB CArbohydrate REsidue check web service. |
 | Customisation and validation of an Oracle AERS Implementation (Oracle Adverse Event Reporting System) |
2010, Thesis
Business processes of a pharmacovigilance department of a major global CRO have been analysed and documented in order to accomplish performance qualification of an Oracle AERS implementation.
|
 | igvCenter: Visualisation of transcription factor binding sites with IGV |
2010, M. Haub, O. M. Nickel
The tool allows browsing and mining for transcription factor binding sites. ChIP-Seq results can be imported and compared with published data.
Some more information can be found on the project page. |































